Abstract
Background Mutations in isocitrate dehydrogenases (IDH) are frequently identified in 10% - 20% of acute myeloid leukemia (AML) cases. IDH mutations induce a neomorphic enzymatic function that catalyzes the conversion of α-ketoglutarate (α-KG) to the R enantiomer of 2-hydroxyglutarate (R-2HG). α-KG is also a co-substrate, together with oxygen and the co-factor iron, for α-KG-dependent dioxygenases. R-2HG is structurally close to α-KG and competitively inhibits the activity of α-KG-dependent dioxygenases. It has been reported that R-2HG inhibits the growth of IDH-wild-type AML cells by inhibiting the activity of fat mass and obesity-associated protein (FTO) and then reducing MYC and CEBPA expression. Recently, R-2HG was found to suppress aerobic glycolysis and proliferation in myeloid leukemia cells. However, why AML cells carrying IDH mutations tolerated high levels of R-2HG is not clear.
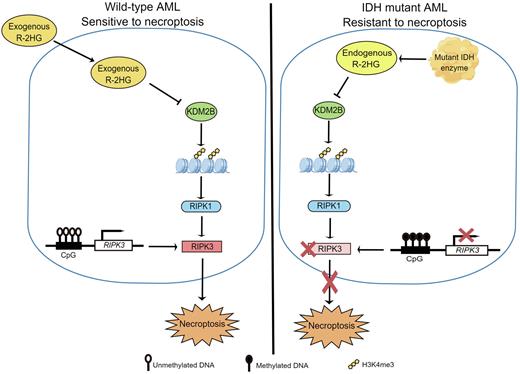
Results In line with the previous study, we found that R-2HG markedly inhibited the viability of AML cells in a dose- and time-dependent manner. Interestingly, Necrostatin-1(Nec-1), a necroptosis inhibitor, could significantly prevent cell death induced by R-2HG in AML cell lines, but not Ferrostatin-1 (a ferroptosis inhibitor), Z-VAD (an apoptosis inhibitor), 3-MA (an autophagy inhibitor) and VX-765 (a pyroptosis inhibitor), suggesting that necroptosis contributed to the anti-leukemia activity of R-2HG.
We evaluated the expression of genes involved in the necroptosis pathway and found the level of RIPK1 was significantly increased in R-2HG treated SKM-1 and THP-1 cells, while the expressions of RIPK3 and MLKL did not change. The knockdown of RIPK1 reduced the necroptosis caused by R-2HG in SKM-1 and THP-1 cells. Besides, a significant decrease in proliferation was observed when RIPK1 was overexpressed in AML cells which was significantly rescued by Nec-1.
Among α-KG-dependent dioxygenases, the expression of lysine demethylase 2B (KDM2B) was negatively correlated with that of RIPK1. Moreover, KDM2B deficiency significantly increased RIPK1 expression and triggered necroptosis. ChIP followed by qPCR (ChIP-qPCR) showed that the RIPK1 promoter region was significantly enriched by H3K4me3, a main target of KDM2B. R-2HG treatment or knockdown of KDM2B activated H3K4me3 and increased RIPK1 expression.
By comparing the expression of necroptosis-related genes between R-2HG-resistant and sensitive cell lines (GSE87187), we found that RIPK3, another key player of necroptosis, was under-expressed in R-2HG-resistant cell lines. Consistently, RIPK3 was lowly expressed in IDH-mutant primary AML cells compared to that of IDH wild-type primary AML cells. We found that enhanced RIPK3 levels increased the toxicity of R-2HG in R-2HG-resistant cell lines. Methylation-specific PCR (MS-PCR) revealed that methylation occurred in this region of genomic DNA near the TSS of RIPK3 in R-2HG-resistant cell lines. Primary cells from IDH-mutant AML patients were more sensitive to decitabine than those from IDH wild-type AML patients.
Conclusion Herein, we reported that R-2HG triggered necroptosis in AML cell lines and IDH wild-type primary AML cells via KDM2B/RIPK1 signal pathway. The loss of RIPK3 expression resulting from RIPK3 promoter hypermethylation contributed to R-2HG tolerance in IDH mutated primary AML cells and R-2HG-resistant AML cell lines. IDH-mutant AML cells were more sensitive to DNMT inhibitor. These results implicate that restoring RIPK3 activity is a promising way to reverse R-2HG-resistant in AML cells with IDH mutation.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal